While the COVID-19 vaccines introduced many people to RNA-based medicines, RNA oligonucleotides have already been on the market for years to treat diseases like Duchenne Muscular Dystrophy and amyloidosis. RNA therapies offer many advantages over traditional small molecule drugs, including their ability to address almost any genetic component within cells and to guide gene editing tools like CRISPR to their targets.
However, the promise of RNA is currently limited by the fact that rapidly growing global demand is outpacing the industry’s ability to manufacture it. The standard method of chemically synthesizing RNA was invented in the 1980s, and requires specialized equipment and labor-intensive processes.
Chemical synthesis methods are also limited in terms of the range of nucleotide building blocks that they can incorporate into RNA molecules, and they produce metric tons of toxic chemical byproducts that create environmental hazards and limit factories’ production capacity. These problems will only increase as RNA production ramps up in response to demand.
A team of scientists at the Wyss Institute at Harvard University and Harvard Medical School (HMS) has created a solution to this problem: a new RNA synthesis process that expands the RNA therapeutic design space and unlocks the potential for rapid scale-up that chemical synthesis cannot achieve.
Their novel method can produce RNA with efficiencies and purities comparable to current industry standards using water and enzymes rather than the toxic solvents and explosive catalysts that plague current manufacturing. It can also incorporate all the common molecular modifications that are found in RNA drugs today, and has the potential to incorporate novel RNA chemistries for new types of therapies.
The achievement is described in a paper published today in Nature Biotechnology.
“As demand for RNA drugs continues to grow and additional products come to market, we will exceed the current global supply of acetonitrile, the organic solvent used in chemical RNA synthesis methods,” said co-first author Jonathan Rittichier, Ph.D., a former Postdoctoral Fellow at the Wyss and HMS. He and fellow first author and former Wyss Research Scientist Daniel Wiegand, M.S.Ch.E.; Wyss Core Faculty member George Church, Ph.D., and others co-founded EnPlusOne Biosciences to commercialize their technology.
“Delivering RNA drugs to the world at these scales requires a paradigm shift to a renewable, aqueous synthesis, and we believe our proprietary enzymatic technology will enable that shift,” Rittichier added.
A better, bio-friendly way
In Church’s lab, Rittichier, Wiegand and co-corresponding author Erkin Kuru, Ph.D. recognized that the pharmaceutical industry was in the midst of an RNA revolution. The lab had previously devised a way to synthesize DNA using enzymes, and hypothesized they could do the same for RNA.
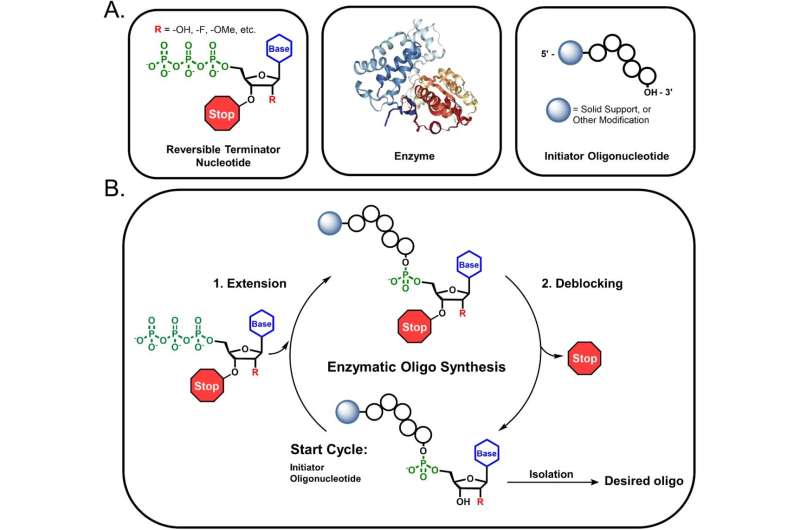
The scientists started with an enzyme from a strain of yeast, Schizosaccharomyces pombe, which is known to link nucleotide molecules together to form strands of RNA. They engineered the enzyme to make it more efficient and capable of incorporating non-standard nucleotides into RNA. This was especially important in building a useful drug development platform, as every FDA-approved RNA drug contains nucleotides that have been modified from their original form to increase their stability in the body or endow them with new functions.
They then focused on the nucleotides themselves. In standard chemical RNA synthesis, nucleotides have “protecting groups” added to them: a sort of chemical bubble-wrap that prevents the molecule from being damaged by the harsh reaction conditions.
These protecting groups must be removed after synthesis in order for the RNA to function, and this process requires an additional round of chemical reactions that can damage the RNA as it’s being built. The milder conditions of EnPlusOne’s synthesis eliminate the need for any bulky bubble-wrap, ultimately leading to better manufacturing.
But even as it solved one problem, the team’s enzyme introduced a different one: Its natural activity would string together nucleotides uncontrollably, resulting in inaccurate RNA sequences. To solve this issue, they modified their nucleotides with a “blocker,” a chemical group that stalls the enzyme and only allows for the addition of one nucleotide at a time. Once the desired nucleotide has been added, the blocker is removed to allow the next nucleotide in the sequence to bind, resulting in a two-step process that is simpler and less reagent-intensive than the typical four-step chemical synthesis method.
The researchers demonstrated that their new process incorporated nucleotides with 95% efficiency, which is comparable to chemical synthesis. The team then iteratively repeated cycles of enzymatic RNA synthesis to build molecules of 10 nucleotides in length. They are now routinely able to build molecules that are 23 nucleotides long, which is the size of many blockbuster RNA therapeutics.
From molecules to medicines
The key to turning RNA into useful drugs is modifying its naturally existing nucleotides. The team also demonstrated that their enzymatic synthesis method could successfully produce strands of RNA with multiple types of modified nucleotides with the same ability as natural nucleotides.
“Natural RNA is made of four letters—A, U, C, and G—but we can expand this simple alphabet with synthetic biology,” said Kuru, who is a Postdoctoral Fellow at HMS. “Our process essentially increases the number of keys we have on our ‘RNA typewriter’ to a much richer alphabet that we can use to write RNAs with new functions and properties.”
This work formed the basis for a Validation Project at the Wyss Institute in 2019 and 2020, when it was de-risked and prepared for commercialization.
“Enzymatic nucleotide synthesis technologies offer many advantages as an alternative to chemical-based methods. This platform can help unlock the immense potential of RNA therapeutics in a sustainable way, especially manufacturing high-quality guide RNA molecules for CRISPR/Cas gene editing,” said co-corresponding author Church, who is also the Robert Winthrop Professor of Genetics at HMS.
EnPlusOne is also using its platform to manufacture small interfering RNAs (siRNAs) at lab scale that could be used to treat a wide variety of diseases.
“RNA drugs offer a powerful new treatment approach for a huge range of diseases. However, current manufacturing methods for these drugs are limited in terms of the chemical diversity they can produce, the amount of material that can be produced at a reasonable cost, and their negative impact on the environment due to the harsh chemicals they require. EnPlusOne’s elegant bioinspired enzyme synthesis alternative offers a way to overcome all these limitations, and could help the RNA therapeutics industry to explode,” said Wyss Founding Director Don Ingber, M.D., Ph.D.
Additional authors of the Nature Biotechnology paper include Ella Meyer, Howon Lee, Nicholas J. Conway, Daniel Ahlstedt, Zeynep Yurtsever, and Dominic Rainone. Lee and Ahlstedt are also co-founders of EnPlusOne.
More information:
Template-independent enzymatic synthesis of RNA oligonucleotides, Nature Biotechnology (2024). DOI: 10.1038/s41587-024-02244-w
Citation:
A better way to make RNA drugs: Enzymatic synthesis method expands capabilities while eliminating toxic byproducts (2024, July 12)
retrieved 12 July 2024
from https://phys.org/news/2024-07-rna-drugs-enzymatic-synthesis-method.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
































